Miranda Orr’s Spatial Analyses from Discovery to Validation Reveal Key Processes in Alzheimer’s Disease

Miranda E. Orr, PhD, is a leading voice in neurology and geriatric medicine at Washington University in St. Louis. Dr. Orr and her team of neuroscience experts continue to unravel the complexities of Alzheimer’s disease (AD). Their research addresses how the neuronal, glial, and vascular compartments respond to amyloid β plaques and tau aggregates, looks for additional AD hallmarks, and maps how aging and disease processes differ in subtle but significant ways across various parts of the brain.
To pursue this goal, the Orr lab leverages a suite of spatial biology instruments that collectively provide unprecedented resolution of disease physiology. Optimal for discovery workflows, the GeoMx® Digital Spatial Profiler (DSP) provides the highest plex spatial multiomic analysis available today, with whole transcriptome and 570+ protein panels. The Orr lab utilized the newest GeoMx Discovery Proteome Atlas (DPA), containing over 1200 proteins and available later in 2025, to probe healthy and diseased brain tissues. This region-based analysis identified several new biomarkers potentially linked to a diseased state.
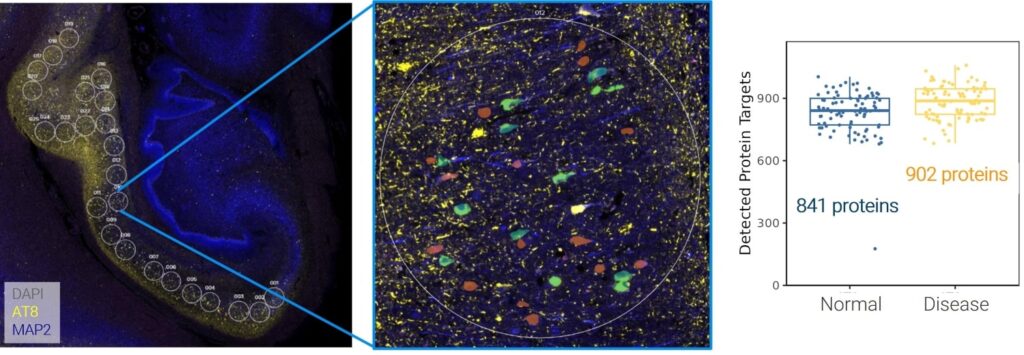
Select areas of interest were chosen for regional analysis of whole transcriptome + over 1000 proteins. High-throughput discovery multiomics enabled the identification of potential AD-associated proteins. These data were presented at the AGBT General Meeting, 2025.
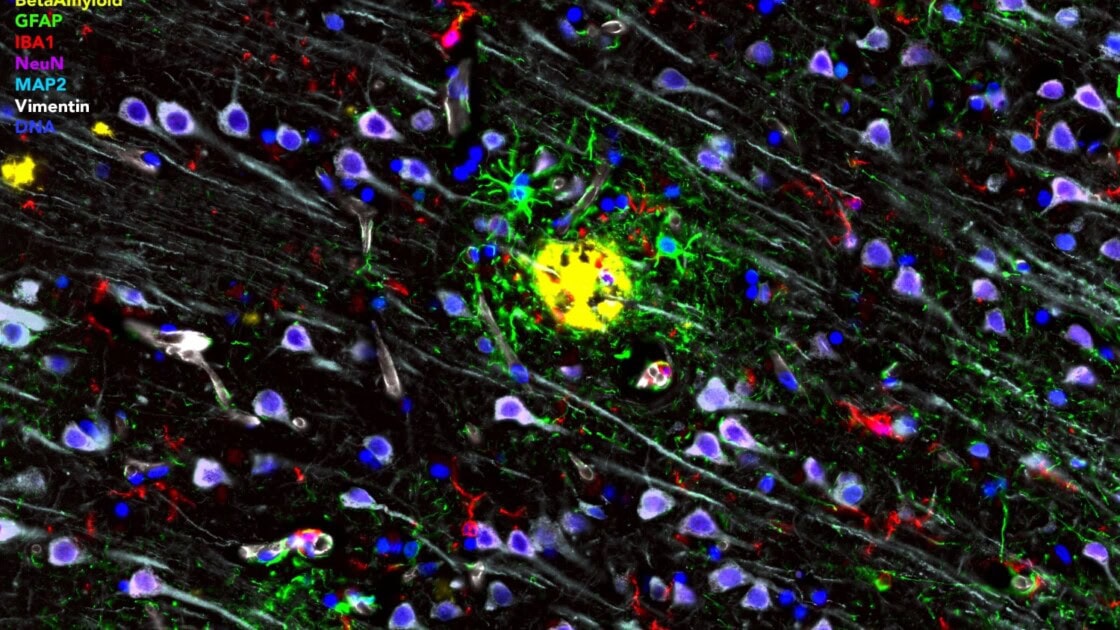
Complementing and validating the regional discovery multiomics perspective of GeoMx DSP, the CellScape™ Precise Spatial Proteomics platform visualizes precise protein biomarker expression of large tissue sections with sub-cellular resolution. In a recent poster presented at the Society for Neuroscience meeting in 2024, Dr. Orr and collaborators showcased the power of the CellScape platform to visualize 20 key proteins simultaneously in formalin-fixed, paraffin-embedded (FFPE) brain samples.
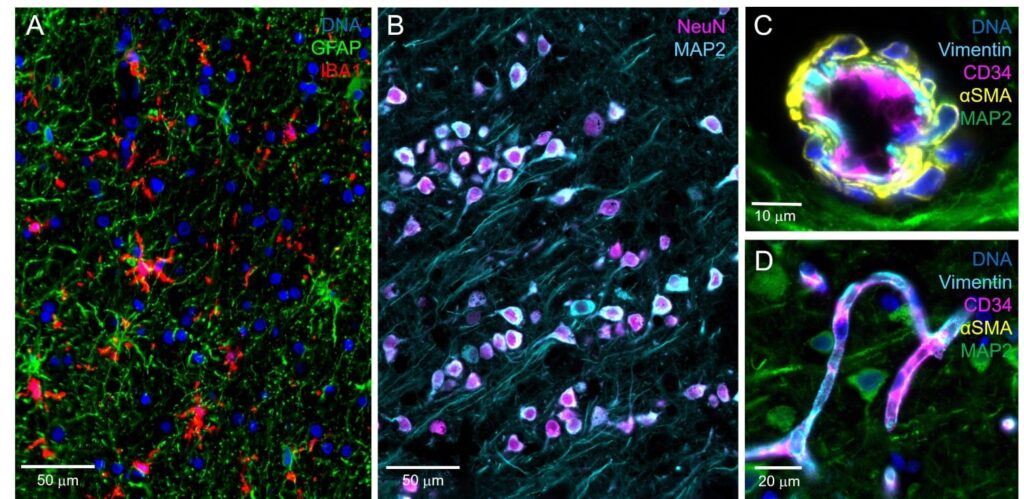
Staining of astroglia and microglia (A), neurons (B), and the cerebral vasculature (C-D) can be achieved with exceptional contrast and clarity at 182 nm/pixel digital sampling.
By iteratively labeling and imaging tissue sections with the CellScape platform, the Orr team visualized neuronal and glial markers, such as MAP2, NeuN, IBA1, and GFAP, alongside pathology-associated proteins including amyloid β, p62, ubiquitin, and multiple phosphorylated tau variants.
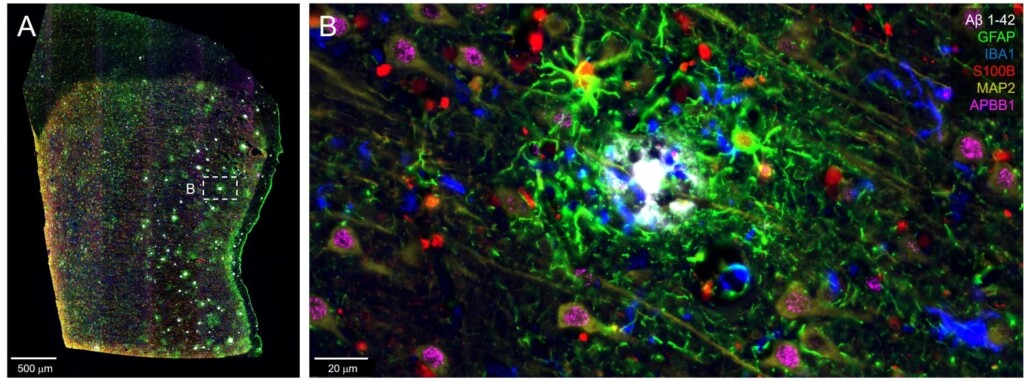
(A) Postcentral gyrus from an AD tissue array shows amyloid β plaques surrounded by reactive astrocytes expressing GFAP, S100B, as well as IBA1+ microglia and MAP2+ neurons (B).
By scanning large tissue sections with the CellScape platform, the Orr lab observed that the AD pathology markers were present in regionally distinct patterns, suggesting that disease progression is not uniform but is instead influenced by local cellular environments. For example, the dentate gyrus region of the hippocampus exhibited sparse tau phosphorylation while neighboring subiculum areas showed robust amyloid β plaque aggregates.
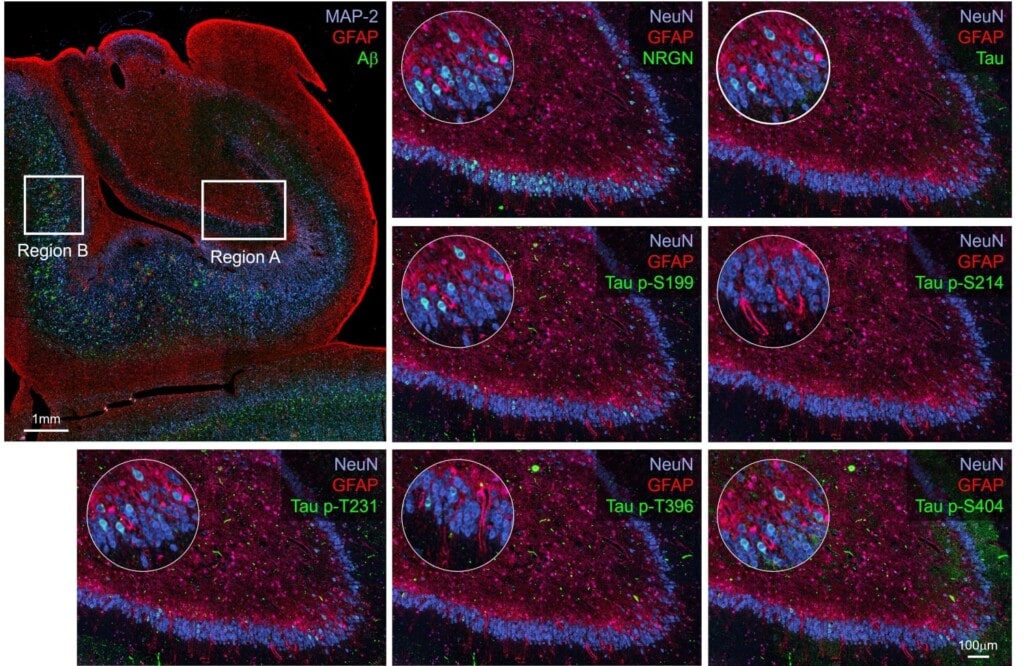
Overview of tissue sample with magnified ROI indicated. Region A: GCL cells express NRGN abundantly in a mosaic pattern. pTAU targets are similarly mosaic in their expression but sparser. For example, Tau p-S199 expressed strongly in some GCL cells, is rather sparse. Cells expressing Tau p-S199 show labeling against other Tau phosphorylation sites as well and only few cells are labeling with only one biomarker. No amyloid β plaques or indications of gliosis were observed in this brain region.
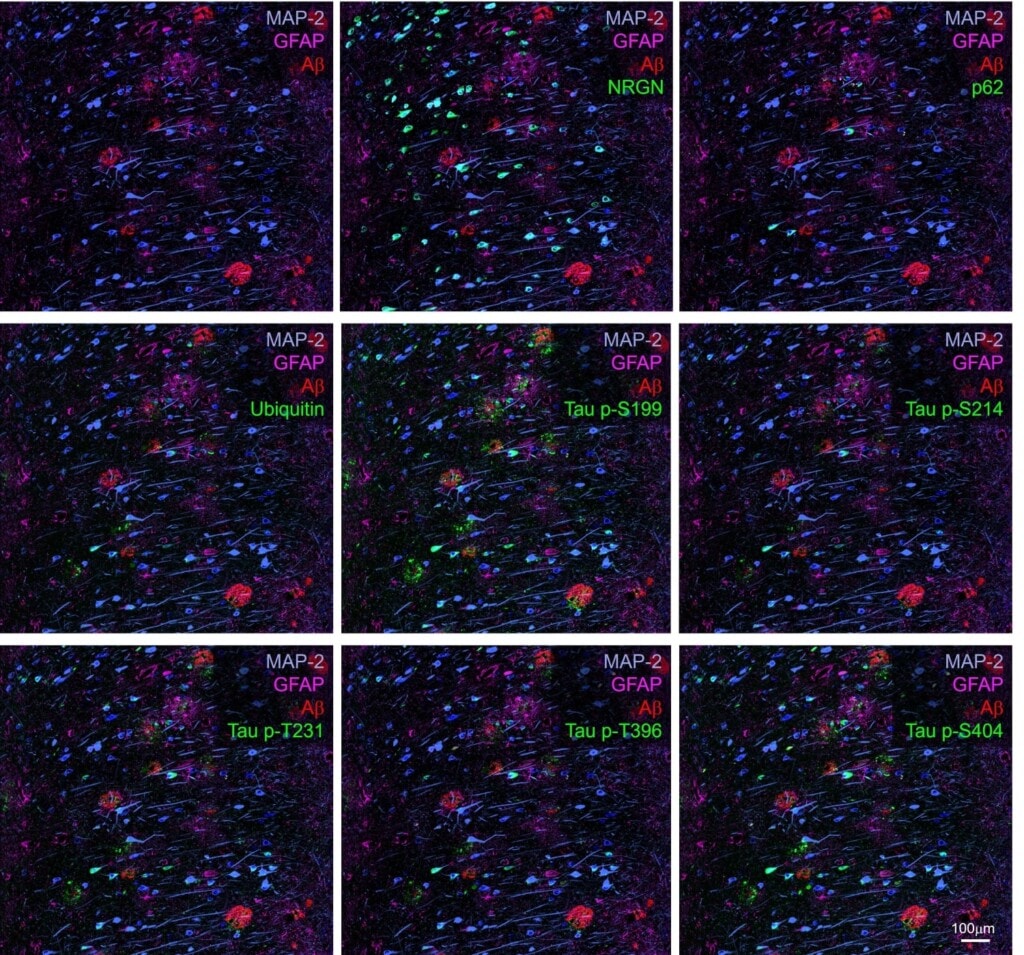
Region B (outlined in previous figure) contained multiple amyloid β plaques of dense and diffuse types. Unlike the GCL cells in the dentate gyrus, neurons in the subiculum labeled comprehensively with the biomarker disease panel. For example, multiple neurons labeled strongly with p62, ubiquitin, Tau p-S214 and Tau p-T396; these markers were not observed in the GCL. These data demonstrate regionally specific labeling for disease biomarkers in Alzheimer’s brain tissues.
By pairing GeoMx DSP regional discovery multiomics with subcellular proteomic imaging using the CellScape platform, the Orr lab can identify new and relevant disease hallmarks and then confirm their presence and map them to specific brain locations. Using these best-in-class spatial technologies together creates a rapid path forward toward understanding disease progression, ultimately aiming toward earlier detection of disease and the creation of targeted and effective treatments.
Miranda Orr’s use of GeoMx DSP, CosMx® Spatial Molecular Imager, and CellScape PSP was also highlighted in a recent Nature feature article – read it now.
Learn more about pairing GeoMx DSP with CellScape Precise Spatial Proteomics by contacting us today.