Why Transcriptomics Matters: Understanding the Value of High-Resolution Spatial Molecular Imaging
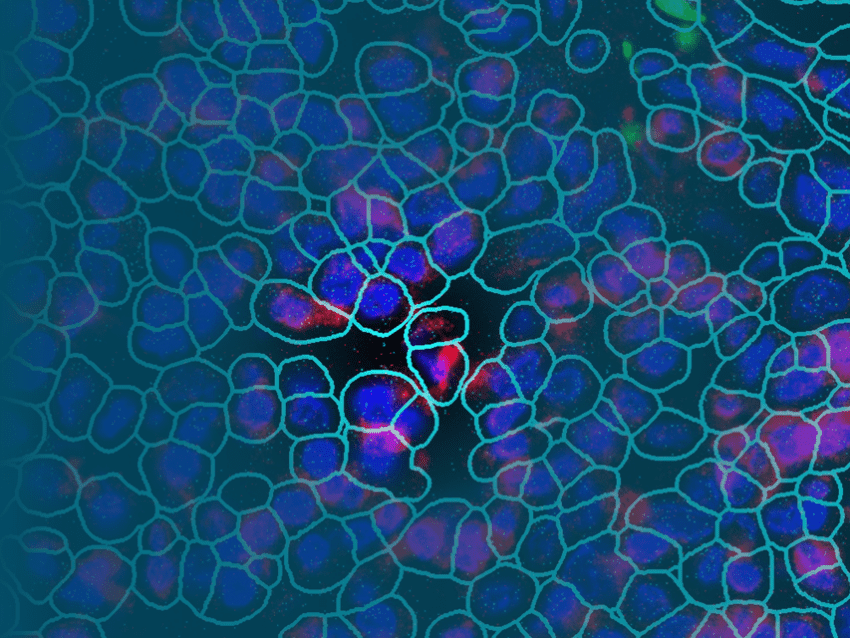
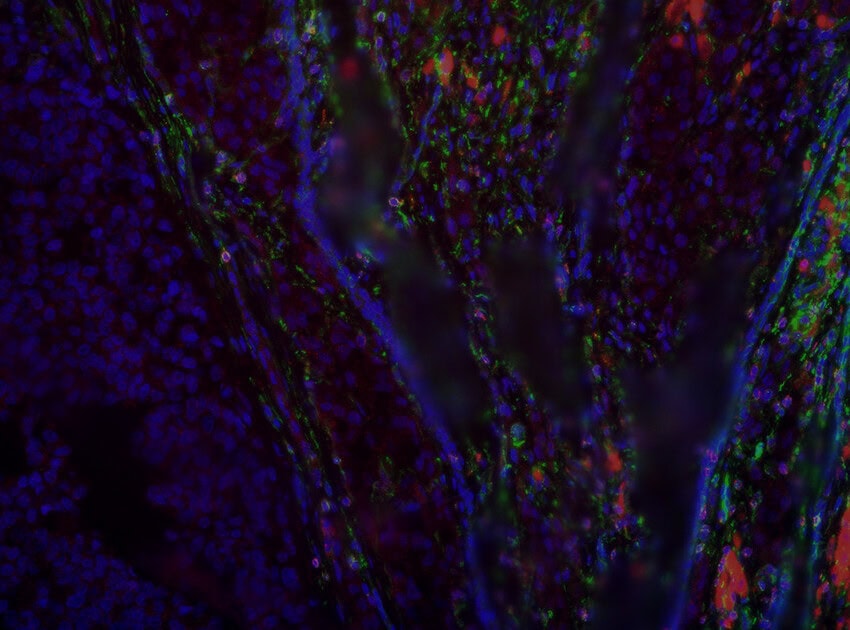
As spatial biology reshapes the future of biomedical research, spatial transcriptomics has emerged as one of the most powerful tools for revealing complex tissue biology. Technologies like the CosMx® Spatial Molecular Imager (SMI) are at the forefront of this transformation, enabling scientists to map gene and protein expression within the native spatial context of intact tissue samples, with a resolution of individual single cells and subcellular structures.
In this article:
- What is Spatial Molecular Imaging?
- What is Spatial Transcriptomics?
- Why Do Researchers Need Tools Like Spatial Molecular Imaging?
- Key Research Areas for Spatial Molecular Imaging
- CosMx vs Xenium: How Spatial Transcriptomics Platforms Compare
- Why Spatial Transcriptomics Is the Future
What Is Spatial Molecular Imaging?
Spatial molecular imaging refers to the visualization of location and abundance of RNA, proteins, or other molecular markers within tissue sections, preserving spatial context. Unlike traditional bulk RNA-sequencing or even single-cell RNA-seq, which require tissue dissociation and lose spatial information, high-plex spatial imaging allows researchers to see exactly where different cell types and gene expression patterns are located within a tissue.
Location matters because biology is inherently spatial. Cell function, communication, and disease progression are all shaped by physical location, microenvironment, and cellular interactions. Whether studying tumor heterogeneity, immune infiltration, or brain architecture, understanding spatial context is critical.
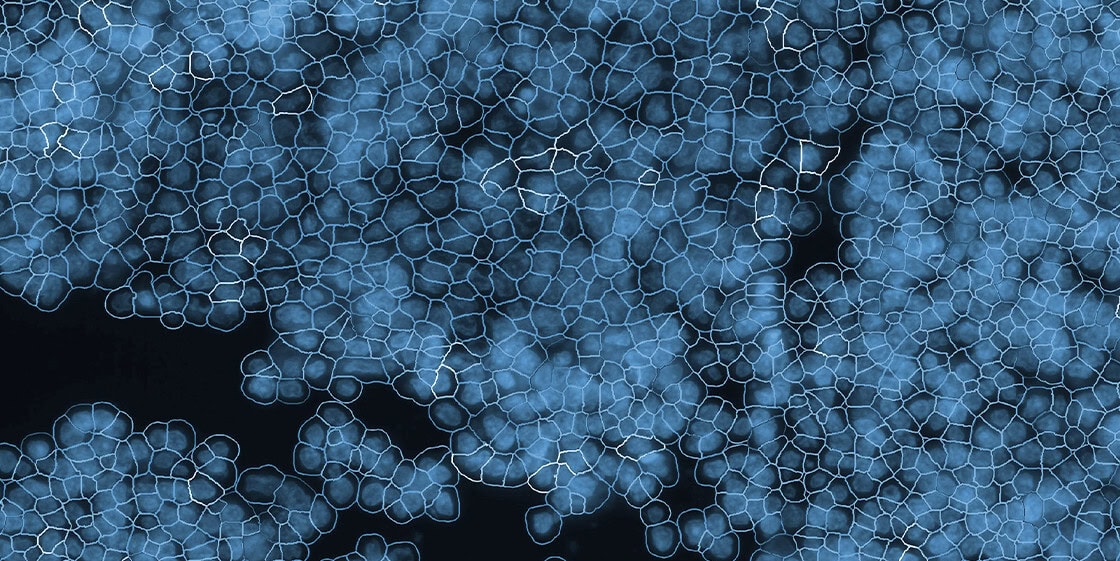
What is Spatial Transcriptomics?
Spatial transcriptomics is the application of spatial molecular imaging to study gene expresion at spatial resolution. It combines in situ hybridization with high-plex detection, capturing hundreds to thousands of RNA transcripts per cell across tissue sections while retaining their precise location coordinates.
This field has rapidly advanced in recent years, with applications across oncology, neuroscience, immunology, infectious disease, and developmental biology. As a result, spatial transcriptomics is now considered a key pillar of multiomics and systems biology approaches.
Read more about spatial transcriptomics.
Why Do Researchers Need Tools Like Spatial Molecular Imaging?
The CosMx Spatial Molecular Imager is purpose-built for high-plex, spatial single-cell analysis of RNA and proteins. The high-fidelity capabilities of the CosMx platform stem from its unmatched sensitivity, highest single-cell plex, and accurate cell segmentation, which support the most critical needs of spatial transcriptomics researchers:
- Subcellular resolution: CosMx SMI enables the identification of cell types, states, and interactions at a level not possible with lower-resolution methods, resolving transcripts within the nucleus, cytoplasm, or membrane.
- High sensitivity: The CosMx platform detects more transcripts per cell than competing platforms, without sacrificing specificity or resolution, enabling robust analysis even in low-expressing or degraded samples [1].
- High-plex same-cell multiomics: Researchers can measure thousands of RNA molecules and even the entire human transcriptome plus 72 proteins within a single FFPE or fresh frozen (FF) tissue section with CosMx Same-Cell Multiomics.
- Direct, amplification-free RNA detection: Like the nCounter® Analysis Platform, CosMx SMI uses direct hybridization to detect RNA without enzymatic amplification, preserving true transcript abundance. Amplification-based methods can introduce bias, distorting expression levels, especially in degraded or low-input samples [2].
- True spatial context: With no need to dissociate tissue, CosMx SMI preserves tissue architecture, cell morphology, and microenvironment structure, which are critical for studying tumors, immune infiltration, and complex neural tissue.
- Compatibility with fixed samples: The CosMx workflow supports both FFPE and FF tissue, enabling spatial transcriptomics in archival, biobanked specimens and translational research settings.
- Accurate cell segmentation: Data analysis for CosMx SMI can be completed using the cloud-based AtoMx® Spatial Informatics Platform (SIP), which leverages best-in-class AI algorithms for the most accurate definition of cell boundaries and assignment of transcripts to individual cells.

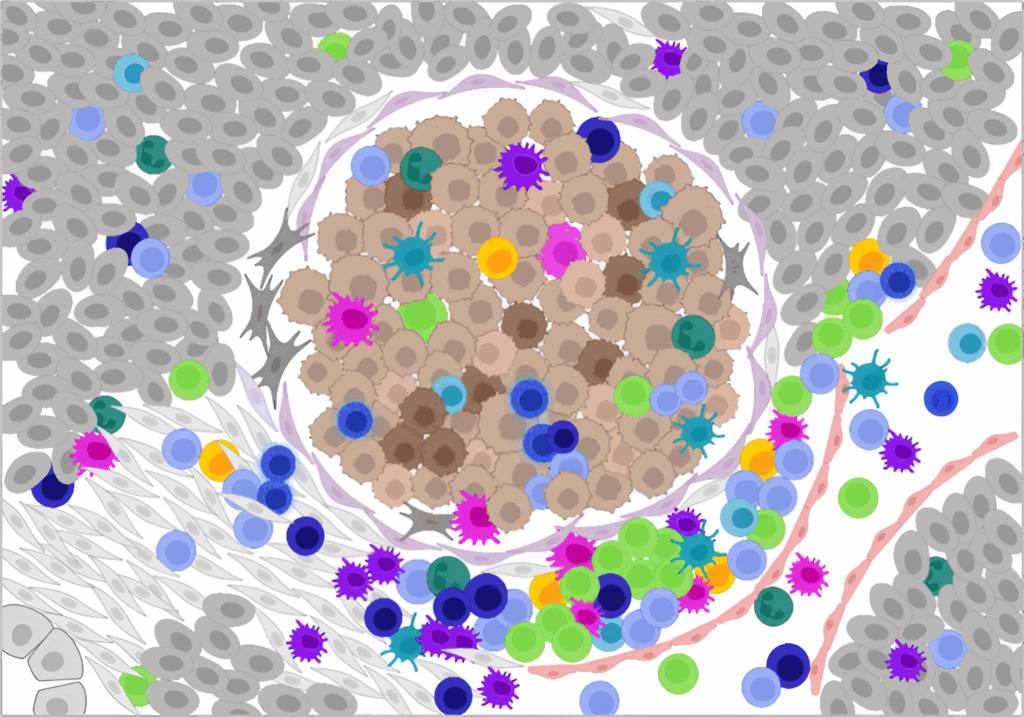
Key Research Areas for Spatial Molecular Imaging
The CosMx SMI is being used across a range of biological systems and disease models to answer questions that are inaccessible with dissociated cell or bulk profiling approaches. Whole transcriptome spatial analysis at subcellular resolution now allows researchers in a variety of fields to profile gene and protein expression across intact tissues while preserving spatial
context:
- Tumor microenvironment profiling: Map spatial interactions between cancer cells, immune infiltrates, and stroma to understand treatment resistance and response.
- Neuroscience: Resolve neuronal and glial subtypes in dense brain tissue and identify spatial gene expression patterns involved in neurodevelopment, neuroinflammation, and neurodegeneration.
- Immunology and infectious disease: Track immune cell dynamics and pathogen-host interactions at cellular resolution.
- Tissue atlasing: Build pathways-first maps of healthy and diseased tissues using whole transcriptome analysis.
CosMx vs Xenium: How Spatial Transcriptomics Platforms Compare
Multiple platforms have emerged to meet growing demand for high-plex, high-resolution spatial profiling. Among the most discussed are the CosMx SMI from Bruker Spatial Biology and the Xenium In Situ platform from 10x Genomics. Both are classified as single-cell spatial transcriptomics platforms, but their capabilities diverge in meaningful ways.
1. Sensitivity: Setting the Record Straight
CosMx SMI consistently demonstrates greater transcript detection sensitivity across a wide range of tissue types and panels than the Xenium platform. In several well-designed independent studies, the CosMx platform detected more transcripts per cell on average than Xenium, particularly when using the CosMx 6K RNA panel [1]. These findings reflect key differences in panel design, probe chemistry, background suppression algorithms, and cell segmentation in data analysis pipelines [3].
Importantly, CosMx SMI does not compromise sensitivity for plex size. Researchers get more transcripts, across more targets, with the spatial resolution needed to distinguish neighboring cells and subcellular compartments. Learn More.
2. Resolution and Cellular Segmentation
While many platforms offer single-cell resolution, CosMx SMI provides true subcellular imaging of RNA using antibody-based segmentation markers. This allows researchers to localize transcripts to the nucleus, cytoplasm, or membrane, and to resolve tightly packed or irregularly shaped cells, which can be critical for analysis of tissues like brain, tumor, and lymphoid organs.
Additionally, CosMx SMI users can tune cell segmentation parameters in AtoMx SIP, which allows further refinement of cell segmentation results and enables accurate attribution of every
transcript to its correct cell [4].
3. Plex Capacity
CosMx SMI supports whole transcriptome spatial analysis with the Whole Human Transcriptome panel (WTX) comprised of approximately 19,000 RNA targets, enabling unbiased discovery across the entire transcriptome. The Xenium platform is currently limited to a maximum of 5,000 RNA targets [5].
4. Protein + RNA Co-detection in the Same Cell
CosMx SMI enables simultaneous spatial analysis of the whole human transcriptome and key proteins for powerful same-cell multiomics. CosMx protein assays use panels of validated, oligo-conjugated antibodies and allow researchers to measure up to 76 protein targets on the same tissue section used for RNA analysis. This co-detection capability enables high-resolution, multimodal phenotyping of complex tissues, metabolic pathway activity, and cell states. This level of detail is not achievable using lower plex assays from competitor platforms.
5. Final Takeaway: CosMx Leads in Performance
While the CosMx, Xenium, and other platforms fall under the umbrella of single-cell spatial transcriptomics, CosMx stands apart in terms of sensitivity, resolution, plex, and same-cell multiomics capability. With the highest transcript detection, true subcellular resolution, whole transcriptome coverage, and integrated high-plex same-cell multiomics, CosMx delivers a comprehensive and biologically meaningful view of tissue architecture.
Why Spatial Transcriptomics Is the Future
Spatial transcriptomics offers a powerful way to study gene and protein expression within intact tissue architecture. With high sensitivity and exploration of the entire human whole transcriptome at subcellular resolution as well as support for both RNA and protein detection, the CosMx SMI platform allows researchers to characterize complex cell types, states, and interactions that are difficult to resolve using dissociated or bulk profiling methods.
Today, spatial molecular imaging is enabling new discoveries across oncology, neuroscience, immunology, and other fields where spatial context matters. By preserving the relationships between cells and their microenvironments, researchers can better understand the biology driving development, disease, and therapeutic response.
References
1. CosMx SMI vs Xenium: Superior in situ single-cell performance study
2. Biases in RNA-seq Library Preparation: Current challenges and solutions
3. CosMx SMI vs Xenium: Comparative analysis of breast tissue
4. A complete pipeline for high-plex spatial proteomic profiling and analysis of neural cell
phenotypes on the CosMx Spatial Molecular Imager and AtoMx Spatial Informatics
Platform
5. Aiming high: 10x, NanoString Launch Large-Scale Panels